Introduction
FRAP (Fluorescence Recovery After Photobleaching) is a method for the diffusion kinetics (and also, correctly, reaction-diffusion kinetics) measurements in living cells using fluorescence microscopy (Zlatanov, 1987; Lopez, 1988; Swaisgood, 1989; Anders, 1990; Nagy, 1990; Bryers, 1998; Salomé, 1998; Tinland, 1998; Higgs, 2000; Reits, 2001; Carrero, 2003; Cha 2004; Joubert, 2004; Fukano, 2004; James, 2004; Sprague, 2005; Lele, 2006; Abbaci, 2007; Febo-Ayala, 2007; Travascio, 2007; Dushek, 2008; Kang, 2008; McNally, 2008; Tolentino, 2008; Lambert, 2009; Travascio, 2009; Hagman, 2010; Mueller, 2010; Mai, 2011; Wachsmuth, 2014; Yapp, 2016) which allows to estimate quantitatively the two dimensional lateral diffusion in molecularly thin film containing fluorescent-labeled probes, or for single cell examination (i.e. the study of lateral mobility of cellular molecules). Similar, though less common techniques (also referred to as FRAP) have been developed for the three dimensional diffusion and molecular binding investigation inside the cell. Modern confocal scanning laser microscopes (CSLMs) (Hardingham, 2000; Gribbon, 2001; Loerke,2005; Seiffert, 2005; Braeckmans, 2007; Campbell, 2007; Maung, 2007; Mazza, 2008; Tsibidis, 2008; Kang, 2010; Roberti, 2011; Kang, 2012; De Los Santos, 2015; Lemcke, 2016) are equipped with the system for local photobleaching in selected regions (Figure 1). But currently there are no completely satisfactory methods for molecular-chemical characterization of diffusion / binding kinetics for FRAP using MALDI (Matrix-Assisted Laser Desorption / Ionization) imaging techniques (for example, see Figure 4).

Figure 1. Principle of FRAP. A) The bilayer is uniformly labeled with a fluorescent tag; B) This label is selectively photobleached by a small (~30 μ) fast light pulse; C) The intensity within this bleached area is monitored as the bleached dye diffuses out and new dye diffuses in; D) Eventually uniform intensity is restored. From Wikipedia, the free encyclopedia
[URL: https://en.wikipedia.org/wiki/File:Frap_diagram.svg]
FLIP (Fluorescence Loss in Photobleaching; Figure 2) is a microscopic technique predominantly performed using laser scanning microscopy (e.g. for tagged protein local photobleaching by short, intensive laser excitation on CLSM platform) used for the studies on molecular mobility inside the cells and membranes. The primary application of FLIP is the determination of the continuity of membranous organelles based on the fluorescence intensity in the region of interest and the second less common application of FLIP is the observation of the protein shuttling from the cytoplasm to the nucleus and the determination of the shuttling rate. The above laser-based technique is readily compatible with MALDI (Figure 3) using the same laser beam for MALDI and FLIP and laser spot assisted mapping (Figure 4).
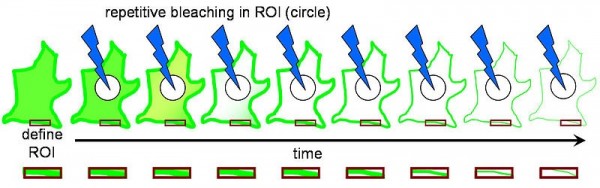
Figure 2. FLIP – decreased fluorescence in a defined region (the red box) adjacent to a bleached region (the circle). From Wikipedia, the free encyclopedia [URL:
https://en.wikipedia.org/wiki/File:Fluorescence_Loss_in_Photobleaching_Schematic.jpg]
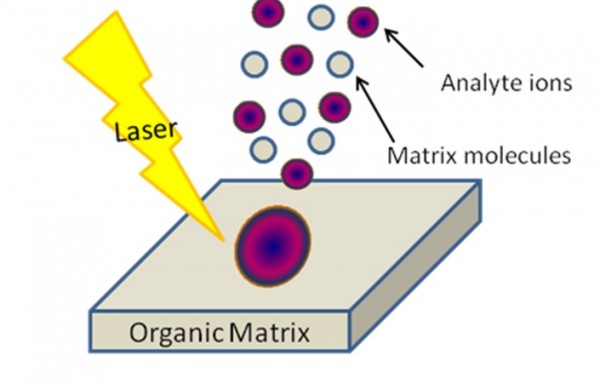
Figure 3. MALDI (Matrix-Assisted Laser Desorption / Ionization). Scheme from MSU tutorial presentation.
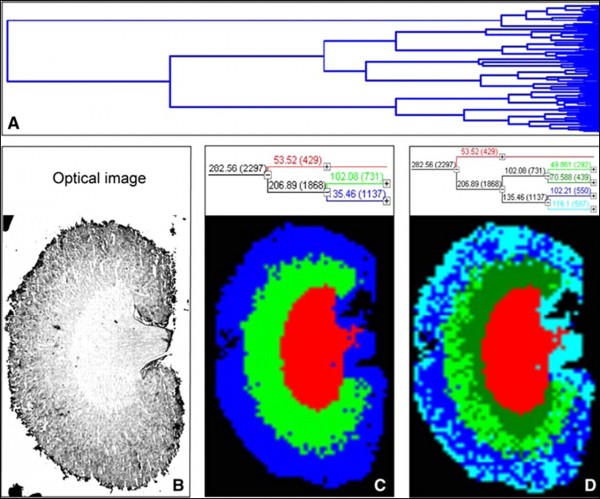
Figure 4. Hierarchical clustering of a mouse kidney data set achieved by MALDI-MSI:
A – full dendrogram of all spectra in a mouse kidney data set. B – optical image of the mouse kidney analyzed by MALDI-MSI. C and D – reconstruction of selected dendrogram branches and corresponding images. The three main branches reflect the renal cortex (blue), medulla (green), and pelvis (red). C, the medulla branch separates into two distinct areas, whereas the cortex branch further differentiates into fat and connective tissue of the renal capsule and hilus and the actual cortex (D). (Walch et al.; from the Archive of Internet)
Methods
In this report we propose to design a new scheme for FRAP measurements based on a coordinate-positioning two-axis stage for MALDI imaging (from typical MALDI heads) and focusable laser sources with programmable microbeam / millibeam positioning. The method is appropriate for standard MALDI heads with nitrogen lasers (337 nm), excimer ArF (193 nm), KrF (248 nm), XeCl (308 nm) and frequency-tripled and quadrupled Nd:YAG lasers (355 nm and 266 nm, respectively). The method can also operate at non-conventional wavelengths, such as 532; 307, 317, 327, 347, and 357 nm (Ahn, 2012), according to the photobleachable carrier spectrum.
Also we propose to design a novel scheme for FLIP measurements using the coordinate-positioning 2D-stage for MALDI imaging (from MALDI head, Feodorov (Notchenko, 2013; Notchenko, 2014; Oganessian, 2014; Gradov, 2015; Gradov, 2015a; Notchenko, 2015) or CLSM stages) and focusable laser sources with microbeam / millibeam positioning. The method is appropriate for standard MALDI heads with nitrogen lasers (337 nm), excimer ArF (193 nm), KrF (248 nm), XeCl (308 nm) and frequency-tripled and quadrupled Nd:YAG lasers (355 nm and 266 nm, respectively). The method can also operate at non-conventional wavelengths if they are compatible with the photobleachable agent spectrum (532; 307, 317, 327, 347, and 357 nm (Ahn, 2012)).
Preliminary data
Many in situ MS-experiments were designed to obtain detailed information on the molecular-chemical basis of fluorescence recovery. Following these studies, computational experiments were carried out which can be used for one-to-one mapping of cytological / histological FRAP patterns and MALDI imaging results with temporal resolution (as well as photobiophysical interactions with soft matter in photobleaching can be correlated with the local MALDI ionization in the same cellular regions). Multi-wavelength hybrid MALDI imaging + MS-FRAP setups can be used for molecular biochemical co-localization studies in hyperspectral (or spectrozonal) imaging regimes. Interactions in far ultraviolet, vacuum ultraviolet and extreme ultraviolet bands may be partially simulated using Geant4 (CERN) in the frames of Geant4-DNA approaches or in terms of particle physics methods (for example: World initStep = laser photon source; World = cellular media; Tank = photo-bleachable vacuole; OutOfWorld = MALDI background target (or wafer); Target = local microscopic FRAP zone) (McCormick, 2003; Rannou, 2004; Dollan, 2005; Lo Meo, 2009; Meo, 2009; Boschi, 2011; Grevillot 2011Blake, 2013; Dooraghi, 2013; Glaser, 2013; Joshirao, 2013; Cuplov, 2014; Nozka, 2014; Tendeiro, 2014; Nilsson, 2015; Pibeleht, 2015; Ogawara, 2016).
The above simulations based on Monte-Carlo methods can be also performed using FLUKA particle transport codes under FORTRAN (in contrast to Geant4 based on C++), but with the known physical / technical and algorithmic limitations. We cannot give a complete list of our performed and planned works due to the limited size of this abstract, but the complex model computational and technical considerations prove our preliminary conclusions about the prospects of hybridization between FRAP and MALDI imaging approaches in the framework of standard serial manufactured MALDI-MS setups.
Conclusion
Since the shuttling rate of a cytoplasm component is directly proportional to its fluorescence decay rate, and if the transport from the nucleus to the cytoplasm occurs, photobleaching will occur in the nucleus as well, the exchange rate between the nucleus and the cytoplasm can be determined from this data. We have developed a novel mathematical apparatus (together with O. Gradow ex-group (Gradov, 2014; Grädow, 2014; Alexandrov, 2015; Gradov, 2015; Jablokow, 2015; Jablokow, 2015a)) required for the investigation of the FLIP molecular mechanisms using kinetic MS studies (including oscillatory and autowave reaction-diffusion regimes), based on temporal resolution and adaptive spatiotemporal resolution of MALDI and MALDI imaging, respectively. Also we have proposed to use the conveyor system for serial slice analyzing (after the automatic microtome / vibratome sectioning module) for MALDI-FLIP-MS-based screening of different tissues on discretely shifting MALDI sample carriers (or a “sample lent” in this case) unwinding from the reel / spool. Thus, several new methods of analysis may be proposed from hybridization of MALDI and FLIP approaches.
References
1. Abbaci M, Barberi-Heyob M, Stines JR, Blondel W, Dumas D, Guillemin F, Didelon J. Gap junctional intercellular communication capacity by gap-FRAP technique: a comparative study. Biotechnol J 2007, 2(1):50-61. DOI: 10.1002/biot.200600092
2. Ahn S-H, Bae Y-J, Kim M-S. Matrix-Assisted Variable Wavelength Laser Desorption Ionization of Peptides; Influence of the Matrix Absorption Coefficient on Expansion Cooling. Bull Korean Chem Soc 2012, 33(9):2955-2960. DOI: 10.5012/bkcs.2012.33.9.2955
3. Alexandrov P , Notchenko A , Gradova M, Gradov O. Simultaneous in situ detection of the optical fluorescence, fluorescence recovery kinetics after photobleaching & membrane ion flux on the electrophysiological lab-on-a-chip. American Journal of Optics and Photonics 2015, 3(5):118-122. DOI: 10.11648/j.ajop.20150305.19
4. Anders JJ, Niedermair S, Ellis E, Salopek M. Response of rat cerebral cortical astrocytes to freeze- or cobalt-induced injury: an immunocytochemical and gap-FRAP study. Glia 1990, 3(6):476-486. DOI: 10.1002/glia.440030606
5. Blake SJ, Vial P, Holloway L, Greer PB, McNamara AL, Kuncic Z. Characterization of optical transport effects on EPID dosimetry using Geant4. Med Phys 2013, 40(4):041708. DOI: 10.1118/1.4794479
6. Boschi F, Meo SL, Rossi PL, Calandrino R, Sbarbati A, Spinelli AE. Optical imaging of alpha emitters: simulations, phantom, and in vivo results. J Biomed Opt 2011, 16(12):126011. DOI: 10.1117/1.3663441
7. Braeckmans K, Remaut K, Vandenbroucke RE, Lucas B, De Smedt SC, Demeester J. Line FRAP with the confocal laser scanning microscope for diffusion measurements in small regions of 3-D samples. Biophys J 2007, 92(6):2172-2183. DOI: 10.1529/biophysj.106.099838
8. Bryers JD, Drummond F. Local macromolecule diffusion coefficients in structurally non-uniform bacterial biofilms using fluorescence recovery after photobleaching (FRAP). Biotechnol Bioeng 1998, 60(4):462-473. DOI: 10.1.1.472.3782
9. Campbell JJ, Knight MM. An improved confocal FRAP technique for the measurement of long-term actin dynamics in individual stress fibers. Microsc Res Tech 2007, 70(12):1034-1040. DOI: 10.1002/jemt.20513
10. Carrero G, McDonald D, Crawford E, de Vries G, Hendzel MJ. Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods 2003, 29(1):14-28. DOI: 10.1016/S1046-2023(02)00288-8
11. Cha B, Kenworthy A, Murtazina R, Donowitz M. The lateral mobility of NHE3 on the apical membrane of renal epithelial OK cells is limited by the PDZ domain proteins NHERF1/2, but is dependent on an intact actin cytoskeleton as determined by FRAP. J Cell Sci 2004, 117(Pt.15):3353-3365. DOI: 10.1242/jcs.01180
12. Cuplov V, Buvat I, Pain F, Jan S. Extension of the GATE Monte-Carlo simulation package to model bioluminescence and fluorescence imaging. J Biomed Opt 2014, 19(2):026004. DOI: 10.1117/1.JBO.19.2.026004
13. De Los Santos C, Chang CW, Mycek MA, Cardullo RA. FRAP, FLIM, and FRET: Detection and analysis of cellular dynamics on a molecular scale using fluorescence microscopy. Mol Reprod Dev 2015, 82(7-8):587-604. DOI: 10.1002/mrd.22501
14. Dollan R. Simulation of optical processes in GEANT4 (DESY Presentation, Feb. 22, 2005). Zeuthen: DESY (Deutsches Elektronen-Synchrotron Ein Forschungszentrum der Helmholtz-Gemeinschaft), from: https://www-zeuthen.desy.de/lcdet/Feb_05_WS/talks/rd_lcdet_sim.pdf
15. Dooraghi AA, Keng PY, Chen S, Javed MR, Kim CJ, Chatziioannou AF, van Dam RM. Optimization of microfluidic PET tracer synthesis with Cerenkov imaging. Analyst 2013, 138(19):5654-5664. DOI: 10.1039/c3an01113e
16. Dushek O, Das R, Coombs D. Analysis of membrane-localized binding kinetics with FRAP. Eur Biophys J 2008, 37(5):627-638. DOI: 10.1007/s00249-008-0286-z
17. Febo-Ayala W, Holland DP, Bradley SA, Thompson DH. Lateral diffusion coefficients of an eicosanyl-based bisglycerophosphocholine determined by PFG-NMR and FRAP. Langmuir 2007, 23(11):6276-6280. DOI: 10.1021/la063720d
18. Fukano T, Hama H, Miyawaki A. Similar diffusibility of membrane proteins across the axon-soma and dendrite-soma boundaries revealed by a novel FRAP technique. J Struct Biol 2004, 147(1):12-18. DOI: 10.1016/j.jsb.2003.11.002
19. Geant4 – User Guide for Application Development: 5.2.5 Optical Photon Processes. Geneve: Conseil Européen pour la Recherche Nucléaire (CERN). from: http://geant4.cern.ch/G4UsersDocuments/UsersGuides/ForApplicationDeveloper/html/TrackingAndPhysics/physicsProcess.html#5.2.5
20. Glaser AK, Kanick SC, Zhang R, Arce P, Pogue BW. A GAMOS plug-in for GEANT4 based Monte Carlo simulation of radiation-induced light transport in biological media. Biomed Opt Express 2013, 4(5):741-759. DOI: 10.1364/BOE.4.000741
21. Gradov O, Gradova M. On the possibility of "MS-patch-clamp" or mass spectrometry hybridization with patch-clamp setups for single cell metabolomics and channelomics. Structure & Function of Biomembranes (International Workshop “Biomembranes’14”), MIPT, 29 September 2014 – 3 October 2014.
22. Gradov OV. Shaking-rotating cultivation neurogoniometry: synchronous technique for gradient cultivation of fish neural tissues and cell cultures on the five-axis mechanized stage and direct time-lapse morphometry of differentiation and proliferation of neural cells. The Cell Cultures of Marine and Freshwater Animals. 2015. Institute of Marine Biology, P.12. DOI: 10.13140/RG.2.1.4297.9682
23. Gradov O, Gradova M. "MS-patch-clamp" or the possibility of mass spectrometry hybridization with patch-clamp setups for single cell metabolomics and channelomics. Advances in Biochemistry 2015, 3:66-71. DOI: 10.11648/j.ab.20150306.11
24. Gradov O, Notchenko A, Oganessian V. The neurogoniometry: Applied optical analysis for neural structure directogramm / radiation pattern measurements. Optics 2015a, 4(6):37-42. DOI: 10.11648.j.optics.20150406.11
25. Grädow O. Novel "phenospectral auxanometry" using complexation of optical spectroscopy and chromatographic auxanometry or GC-MS-auxanometry in forest plant species vegetation phenological monitoring based on gas & flavor chemistry principles. Int J Green Herb Chem. Section A: Green Chem 2014, 3(2):555-579.
26. Grevillot L, Bertrand D, Dessy F, Freud N, Sarrut D. A Monte Carlo pencil beam scanning model for proton treatment plan simulation using GATE/GEANT4. Phys Med Biol 2011, 56(16):5203-5219. DOI: 10.1088/0031-9155/56/16/008
27. Gribbon P, Heng BC, Hardingham TE. Novel confocal-FRAP analysis of carbohydrate-protein interactions within the extracellular matrix. Methods Mol Biol 2001, 171:487-494. DOI: 10.1385/1-59259-209-0:487
28. Hagman J, Lorén N, Hermansson AM. Effect of gelatin gelation kinetics on probe diffusion determined by FRAP and rheology. Biomacromolecules 2010, 11(12):3359-3366. DOI: 10.1021/bm1008487
29. Hardingham T, Gribbon P. Confocal-FRAP analysis of ECM molecular interactions. Methods Mol Biol 2000, 139:83-93. DOI: 10.1385/1-59259-063-2:83
30. Higgs HN. Tools for 'The sceptical Chymist': measuring macromolecular interaction kinetics in live cells by TIR-FRAP. Trends Biochem Sci 2000, 25(11):540-541. DOI: 10.1016/S0968-0004(00)01704-7
31. Interactions of optical photons [Geant 4 – Physics Reference Manual]. Geneve: Conseil Européen pour la Recherche Nucléaire (CERN), from: http://geant4.cern.ch/G4UsersDocuments/UsersGuides/PhysicsReferenceManual/html/node66.html
32. Jablokow A, Gradow O. MS-FRAP or MALDI Imaging Setups With Programmable Laser Sources: a New Way to the Diffusion, Molecular Mobility and Binding Measurements, 63-rd ASMS Conf. on Mass Spectr. & Allied Topics (St. Louis, USA, 2015). DOI: 10.13140/RG.2.1.4919.1841 [see also: JASMS, Vol. 26, Suppl. 1, p. 63 – MP 175; DOI: 10.1007/s13361-015-1158-2].
33. Jablokow A, Gradow O. Verifying Continuity of Membranous Organelles and Measurements of Exchange Rate Between the Nucleus and Cytoplasm using FLIP-Like MALDI-Based Imaging, 63-rd ASMS Conf. on Mass Spectr. & Allied Topics (St. Louis, USA, 2015). DOI: 10.13140/RG.2.1.2322.3203 [see also: JASMS, Vol. 26, Suppl. 1, p. 169 – ThP 662; DOI: 10.1007/s13361-015-1158-2]
34. James PS, Hennessy C, Berge T, Jones R. Compartmentalisation of the sperm plasma membrane: a FRAP, FLIP and SPFI analysis of putative diffusion barriers on the sperm head. J Cell Sci 2004, 117(Pt.26):6485-6495. DOI: 10.1242/jcs.01578
35. Joshirao PM, Shin JW, Vyas CK, Kulkarni AD, Kim H, Kim T, Hong SW, Manchanda VK. Development of optical monitor of alpha radiations based on CR-39. Appl Radiat Isot 2013, 81:184-189. DOI: 10.1016/j.apradiso.2013.06.012
36. Joubert F, Fales HM, Wen H, Combs CA, Balaban RS. NADH enzyme-dependent fluorescence recovery after photobleaching (ED-FRAP): applications to enzyme and mitochondrial reaction kinetics, in vitro. Biophys J 2004, 86(Pt.1):629-645. DOI: 10.1016/S0006-3495(04)74141-7
37. Kang M, Kenworthy AK. A closed-form analytic expression for FRAP formula for the binding diffusion model. Biophys J 2008, 95(2):L13-L15. DOI: 10.1529/biophysj.108.135913
38. Kang M, Day CA, DiBenedetto E, Kenworthy AK. A quantitative approach to analyze binding diffusion kinetics by confocal FRAP. Biophys J 2010, 99(9):2737-2747. DOI: 10.1016/j.bpj.2010.09.013
39. Kang M, Day CA, Kenworthy AK, DiBenedetto E. Simplified equation to extract diffusion coefficients from confocal FRAP data. Traffic 2012, 13(12):1589-1600. DOI: 10.1111/tra.12008
40. Lambert NA. Uncoupling diffusion and binding in FRAP experiments. Nat Methods 2009, 6(3):183-184. DOI:10.1038/nmeth0309-183a
41. Lele TP, Ingber DE. A mathematical model to determine molecular kinetic rate constants under non-steady state conditions using fluorescence recovery after photobleaching (FRAP). Biophys Chem 2006, 120(1):32-35. DOI: 10.1016/j.bpc.2005.10.007
42. Lemcke H, Peukert J, Voronina N, Skorska A, Steinhoff G, David R. Applying 3D-FRAP microscopy to analyse gap junction-dependent shuttling of small antisense RNAs between cardiomyocytes. J Mol Cell Cardiol 2016, 98:117-127. DOI: 10.1016/j.yjmcc.2016.07.008
43. Loerke D, Wienisch M, Kochubey O, Klingauf J. Differential control of clathrin subunit dynamics measured with EW-FRAP microscopy. Traffic 2005, 6(10):918-929. DOI: 10.1111/j.1600-0854.2005.00329.x
44. Lo Meo S, Baldazzi G, Bennati P, Bollini D, Cencelli VO, Cinti MN, Moschini G, Lanconelli N, Navarria FL, Pani R, Pellegrini R, Perrotta A, Vittorini F. Optical physics of scintillation imagers by GEANT4 simulations. Nuclear Instruments and Methods in Physics Research. Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 2009, 607(1): 259-260. DOI:10.1016/j.nima.2009.03.168
45. Lopez A, Dupou L, Altibelli A, Trotard J, Tocanne JF. Fluorescence recovery after photobleaching (FRAP) experiments under conditions of uniform disk illumination. Critical comparison of analytical solutions, and a new mathematical method for calculation of diffusion coefficient D. Biophys J 1988, 53(6):963-970. DOI: 10.1016/S0006-3495(88)83177-1
46. Mai J, Trump S, Ali R, Schiltz RL, Hager G, Hanke T, Lehmann I, Attinger S. Are assumptions about the model type necessary in reaction-diffusion modeling? A FRAP application. Biophys J 2011, 100(5):1178-1188. DOI: 10.1016/j.bpj.2011.01.041
47. Maung NL, Wohland T, Hsu CY. Enamel diffusion modulated by Er:YAG laser. Part 1: FRAP. J Dent 2007, 35(10):787-793. DOI: 10.1016/j.jdent.2007.07.011
48. Mazza D, Braeckmans K, Cella F, Testa I, Vercauteren D, Demeester J, De Smedt SS, Diaspro A. A new FRAP/FRAPa method for three-dimensional diffusion measurements based on multiphoton excitation microscopy. Biophys J 2008, 95(7):3457-3469.
49. McCormick J. Simulating Optical Processes Using Geant4 (NICADD Presentation, 03 Nov. 2003). DeKalb, Illinois: Northern Illinois Center for Accelerator and Detector Development, Northern Illinois University. from: http://nicadd.niu.edu/meetings/dhcal/McCormick_scint_Nov03_2003.ppt
50. McNally JG. Quantitative FRAP in analysis of molecular binding dynamics in vivo. Methods Cell Biol 2008, 85:329-351. DOI: 10.1016/S0091-679X(08)85014-5
51. Meo SL, Bennati P, Cinti MN, Lanconelli N, Navarria FL, Pani R, Pellegrini R, Perrotta A, Vittorini F. A Geant4 simulation code for simulating optical photons in SPECT scintillation detectors. J Instrument 2009, 4(7):P07002. DOI: 10.1088/1748-0221/4/07/P07002
52. Mueller F, Mazza D, Stasevich TJ, McNally JG. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? Curr Opin Cell Biol 2010, 22(3):403-411. DOI: 10.1016/j.ceb.2010.03.002
53. Nagy I, Ohta M, Kitani K. The effect of idebenone on the lateral mobility of proteins in hepatocyte plasma membrane of old rats as revealed by fluorescence recovery after photobleaching (FRAP) technique using an endogenous label. Arch Gerontol Geriatr 1990, 11(3):243-250. DOI: 10.1016/0167-4943(90)90068-H
54. Nilsson J, Cuplov V, Isaksson M. Identifying key surface parameters for optical photon transport in GEANT4/GATE simulations. Appl Radiat Isot 2015, 103:15-24. DOI: 10.1016/j.apradiso.2015.04.017
55. Notchenko AV, Gradov OV. A five-axis arm-manipulator laser system & an algorithm for digital processing of output data for recording and morpho-topological identification of cells and tissue structures. Visualization, Image Processing and Computation in Biomedicine. 2013, 2(1). DOI: 10.1615/VisualizImageProcComputatBiomed.2013005967
56. Notchenko AV, Oganessian VА, Gradov OV. Mechanotronic Neurogoniometry For in vivo & in situ Measurements on Neuroblast Cultures, Neurosphere-Like Stem Cell Clusters, Embryonic Brain Tissues & Living Brain Slices. MIT - Skoltech Biomedical Conference: Towards Therapies of the Future. 2014. DOI: 10.13140/2.1.2616.1281.
57. Notchenko AV, Oganessian VA, Gradov OV. A novel tunable laser diode microrefractometric and goniometric spectrotomography based on multiaxis robotized Feodorov stage and the multi- wavelength spectrorefrctometric technology of the data analysis. MIPT: Proc. II Int. Conf. “PhysTech-Med: Physics of Living Systems”. 2015, P.10-11.
58. Nozka L, Brandt A, Rijssenbeek M, Sykora T, Hoffman T, Griffiths J, Steffens J, Hamal P, Chytka L, Hrabovsky M. Design of Cherenkov bars for the optical part of the time-of-flight detector in Geant4. Opt Express 2014, 22(23):28984-28996. DOI: 10.1364/OE.22.028984
59. Oganessian V, Notchenko A, Gradov O. Mechanotronic Neurogoniometer for in vivo & in situ Measurements on Living Brain Slices and Operating with a Stereotactic Apparatus. Х RGC on Biomedical Engineering. 2014. DOI: 10.13140/2.1.2255.6809.
60. Ogawara R, Ishikawa M. Signal pulse emulation for scintillation detectors using Geant4 Monte Carlo with light tracking simulation. Rev Sci Instrum 2016, 87(7):075114. DOI: 10.1063/1.4959186
61. Pibeleht M. Simulation of optical photons generation and transport [Geant4 –> TREE –> MASTER –> EXAMPLES –> EXTENDED –> OPTICAL] (30 Dec 2015). Lund, Sweden: Lund University; Tallinn, Harjumaa, Estonia: National Institute of Chemical Physics and Biophysics Laboratory of High Energy and Computational Physics. from: https://github.com/mortenpi/geant4/tree/master/examples/extended/optical
62. Rannou FR, Kohli V, Prout DL, Chatziioannou AF. Investigation of OPET Performance Using GATE, a Geant4-Based Simulation Software. IEEE Trans Nucl Sci 2004, 51(5):2713-2717.
63. Reits EA, Neefjes JJ. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat Cell Biol 2001, 3(6):E145-E147. DOI: 10.1038/35078615
64. Roberti MJ, Jovin TM, Jares-Erijman E. Confocal fluorescence anisotropy and FRAP imaging of α-synuclein amyloid aggregates in living cells. PLoS One 2011, 6(8):e23338. DOI: 10.1371/journal.pone.0023338
65. Salomé L, Cazeils JL, Lopez A, Tocanne JF. Characterization of membrane domains by FRAP experiments at variable observation areas. Eur Biophys J 1998, 27(4):391-402. DOI:10.1007/s002490050146
66. Seiffert S, Oppermann W. Systematic evaluation of FRAP experiments performed in a confocal laser scanning microscope. J Microsc 2005, 220(Pt.1):20-30. DOI: 10.1111/j.1365-2818.2005.01512.x
67. Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol 2005, 15(2):84-91. DOI: 10.1016/j.tcb.2004.12.001
68. Swaisgood M, Schindler M. Clonal selection by fluorescence redistribution after photobleaching (FRAP) - a "fast" lateral mobility fibroblast mutant (E7G1). Exp Cell Res 1989, 180(2):529-536. DOI:10.1016/0014-4827(89)90079-7
69. Tendeiro D, Lopes G, Vieira P, Santos JP. Monte Carlo simulation of laser beams interaction with the human eye using Geant4. Biomed Eng Online 2014, 13:58. DOI: 10.1186/1475-925X-13-58
70. Tinland B, Pernodet N, Pluen A. Band broadening in gel electrophoresis: scaling laws for the dispersion coefficient measured by FRAP. Biopolymers 1998, 46(4):201-214. DOI: 10.1002/(SICI)1097-0282(19981005)46:43.0.CO;2-T
71. Tolentino TP, Wu J, Zarnitsyna VI, Fang Y, Dustin ML, Zhu C. Measuring diffusion and binding kinetics by contact area FRAP. Biophys J 2008, 95(2):920-930. DOI: 10.1529/biophysj.107.114447
72. Travascio F, Gu WY. Anisotropic diffusive transport in annulus fibrosus: experimental determination of the diffusion tensor by FRAP technique. Ann Biomed Eng 2007, 35(10):1739-1748. DOU: 10.1007/s10439-007-9346-2
73. Travascio F, Zhao W, Gu WY. Characterization of anisotropic diffusion tensor of solute in tissue by video-FRAP imaging technique. Ann Biomed Eng 2009, 37(4):813-823. DOI: 10.1007/s10439-009-9655-8
74. Tsibidis GD, Ripoll J. Investigation of binding mechanisms of nuclear proteins using confocal scanning laser microscopy and FRAP. J Theor Biol 2008, 253(4):755-768. DOI: 10.1016/j.jtbi.2008.04.010
75. Wachsmuth M. Molecular diffusion and binding analyzed with FRAP. Protoplasma 2014, 251(2):373-382. DOI: 10.1007/s00709-013-0604-x
76. Yapp C, Rogers C, Savitsky P, Philpott M, Müller S. Frapid: achieving full automation of FRAP for chemical probe validation. Biomed Opt Express 2016, 7(2):422-441. DOI: 10.1364/BOE.7.000422
77. Zlatanov IV, Foley M, Birmingham J, Garland PB. Developmental changes in the lateral diffusion of Leydig cell membranes measured by the FRAP method. FEBS Lett 1987, 222(1):47-50. DOI:10.1016/0014-5793(87)80189-8